This post is intended as a quick-start guide to getting a competitive score in the Higgs Boson Machine Learning Challenge, using just a bit of python and scikit-learn.
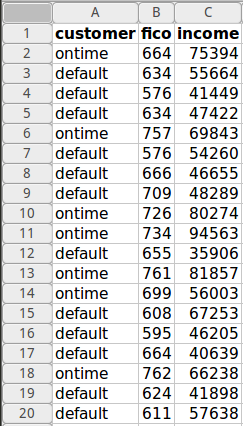
The data consists of just a training.csv and test.csv file. Each of these has a column of ID numbers and 30 columns of feature quantities. Each row represents an event which is either a signal higgs event (s) or a background event (b). The training.csv indicates the truth value (s or b) as well as an event weight.
The goal of this challenge is to classify the events as signal in a manner that optimizes a certain metric. The metric for this challenge is the AMS metric, which is a function of the weighted numbers of correctly and incorrectly guessed signal events.
My approach is pretty simple. I am using the gradient boosting classifier, and I am using the probability output for each event to rank the events. I don’t take the classification at face value – I make a cutoff on the probability prediction and call the upper 15% of events as signal. You can optimize this threshold to maximize the AMS, but after a few tests I have found that the upper 15% is usually about right.
I divide the training sample, keeping 90% as training, and 10% as validation. I calculate the AMS on both to check for overtraining.
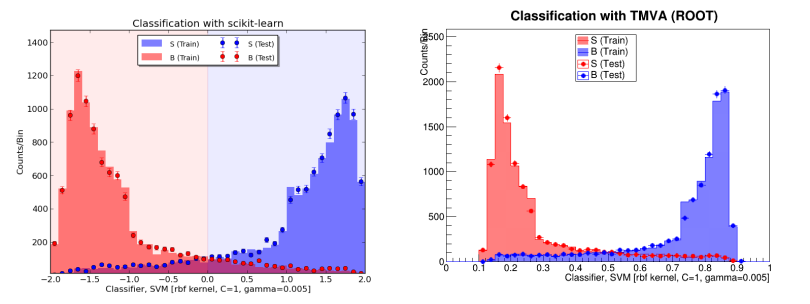
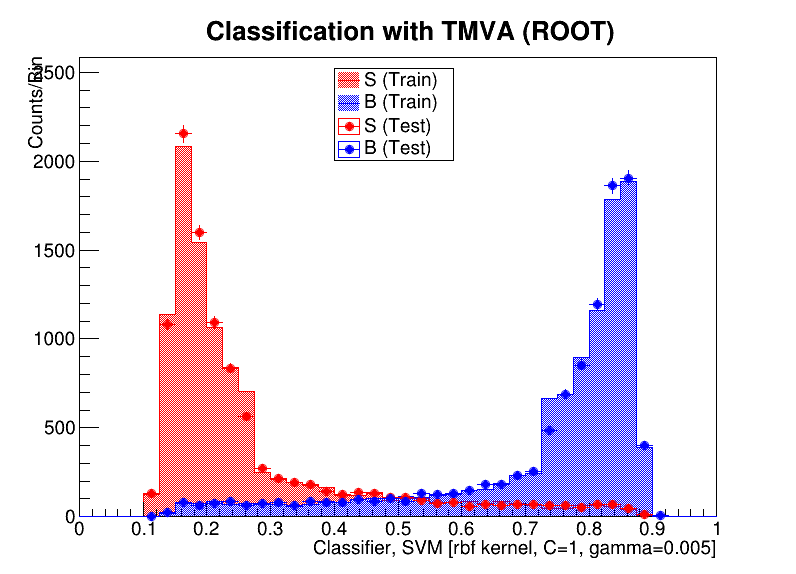
To check that the results look reasonable, I also plot the probability prediction. The red is training background, and the blue is training signal. The black dots are the testing data, normalized to the training sample. I haven’t applied any event weights, because we don’t know these in the test data. The blue shaded region is the upper 15% which I call signal to maximize the AMS metric.
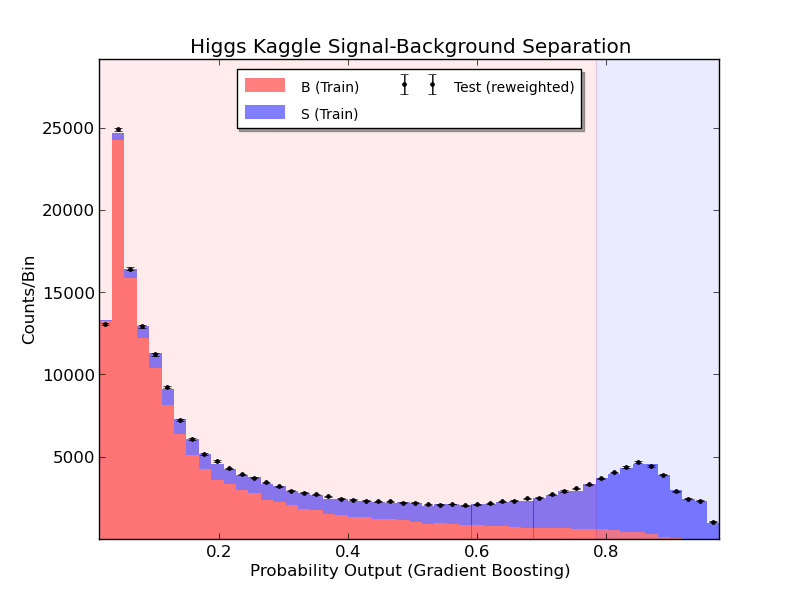
The probability prediction for training signal and background, as well as on testing data, based on the gradient boosting classifier.
Without further ado, this is the code. It should predict in AMS of about 3.47, and the submission scored about 3.38 (either due to random fluctuation or a tiny bit of overtraining). It runs in 5 minutes for me. I am using sklearn 0.14.1 and python 2.7.3.
import numpy as np
from sklearn.ensemble import GradientBoostingClassifier as GBC
import math
# Load training data
print 'Loading training data.'
data_train = np.loadtxt( 'training.csv', delimiter=',', skiprows=1, converters={32: lambda x:int(x=='s'.encode('utf-8')) } )
# Pick a random seed for reproducible results. Choose wisely!
np.random.seed(42)
# Random number for training/validation splitting
r =np.random.rand(data_train.shape[0])
# Put Y(truth), X(data), W(weight), and I(index) into their own arrays
print 'Assigning data to numpy arrays.'
# First 90% are training
Y_train = data_train[:,32][r<0.9]
X_train = data_train[:,1:31][r<0.9]
W_train = data_train[:,31][r<0.9]
# Lirst 10% are validation
Y_valid = data_train[:,32][r>=0.9]
X_valid = data_train[:,1:31][r>=0.9]
W_valid = data_train[:,31][r>=0.9]
# Train the GradientBoostingClassifier using our good features
print 'Training classifier (this may take some time!)'
gbc = GBC(n_estimators=50, max_depth=5,min_samples_leaf=200,max_features=10,verbose=1)
gbc.fit(X_train,Y_train)
# Get the probaility output from the trained method, using the 10% for testing
prob_predict_train = gbc.predict_proba(X_train)[:,1]
prob_predict_valid = gbc.predict_proba(X_valid)[:,1]
# Experience shows me that choosing the top 15% as signal gives a good AMS score.
# This can be optimized though!
pcut = np.percentile(prob_predict_train,85)
# This are the final signal and background predictions
Yhat_train = prob_predict_train > pcut
Yhat_valid = prob_predict_valid > pcut
# To calculate the AMS data, first get the true positives and true negatives
# Scale the weights according to the r cutoff.
TruePositive_train = W_train*(Y_train==1.0)*(1.0/0.9)
TrueNegative_train = W_train*(Y_train==0.0)*(1.0/0.9)
TruePositive_valid = W_valid*(Y_valid==1.0)*(1.0/0.1)
TrueNegative_valid = W_valid*(Y_valid==0.0)*(1.0/0.1)
# s and b for the training
s_train = sum ( TruePositive_train*(Yhat_train==1.0) )
b_train = sum ( TrueNegative_train*(Yhat_train==1.0) )
s_valid = sum ( TruePositive_valid*(Yhat_valid==1.0) )
b_valid = sum ( TrueNegative_valid*(Yhat_valid==1.0) )
# Now calculate the AMS scores
print 'Calculating AMS score for a probability cutoff pcut=',pcut
def AMSScore(s,b): return math.sqrt (2.*( (s + b + 10.)*math.log(1.+s/(b+10.))-s))
print ' - AMS based on 90% training sample:',AMSScore(s_train,b_train)
print ' - AMS based on 10% validation sample:',AMSScore(s_valid,b_valid)
# Now we load the testing data, storing the data (X) and index (I)
print 'Loading testing data'
data_test = np.loadtxt( 'test.csv', delimiter=',', skiprows=1 )
X_test = data_test[:,1:31]
I_test = list(data_test[:,0])
# Get a vector of the probability predictions which will be used for the ranking
print 'Building predictions'
Predictions_test = gbc.predict_proba(X_test)[:,1]
# Assign labels based the best pcut
Label_test = list(Predictions_test>pcut)
Predictions_test =list(Predictions_test)
# Now we get the CSV data, using the probability prediction in place of the ranking
print 'Organizing the prediction results'
resultlist = []
for x in range(len(I_test)):
resultlist.append([int(I_test[x]), Predictions_test[x], 's'*(Label_test[x]==1.0)+'b'*(Label_test[x]==0.0)])
# Sort the result list by the probability prediction
resultlist = sorted(resultlist, key=lambda a_entry: a_entry[1])
# Loop over result list and replace probability prediction with integer ranking
for y in range(len(resultlist)):
resultlist[y][1]=y+1
# Re-sort the result list according to the index
resultlist = sorted(resultlist, key=lambda a_entry: a_entry[0])
# Write the result list data to a csv file
print 'Writing a final csv file Kaggle_higgs_prediction_output.csv'
fcsv = open('Kaggle_higgs_prediction_output.csv','w')
fcsv.write('EventId,RankOrder,Class\n')
for line in resultlist:
theline = str(line[0])+','+str(line[1])+','+line[2]+'\n'
fcsv.write(theline)
fcsv.close()
Lastly, if you are interested in drawing the distribution as I have, this is the code:
from matplotlib import pyplot as plt
Classifier_training_S = gbc.predict_proba(X_train[Y_train>0.5])[:,1].ravel()
Classifier_training_B = gbc.predict_proba(X_train[Y_train<0.5])[:,1].ravel()
Classifier_testing_A = gbc.predict_proba(X_test)[:,1].ravel()
c_max = max([Classifier_training_S.max(),Classifier_training_B.max(),Classifier_testing_A.max()])
c_min = min([Classifier_training_S.min(),Classifier_training_B.min(),Classifier_testing_A.min()])
# Get histograms of the classifiers
Histo_training_S = np.histogram(Classifier_training_S,bins=50,range=(c_min,c_max))
Histo_training_B = np.histogram(Classifier_training_B,bins=50,range=(c_min,c_max))
Histo_testing_A = np.histogram(Classifier_testing_A,bins=50,range=(c_min,c_max))
# Lets get the min/max of the Histograms
AllHistos= [Histo_training_S,Histo_training_B]
h_max = max([histo[0].max() for histo in AllHistos])*1.2
# h_min = max([histo[0].min() for histo in AllHistos])
h_min = 1.0
# Get the histogram properties (binning, widths, centers)
bin_edges = Histo_training_S[1]
bin_centers = ( bin_edges[:-1] + bin_edges[1:] ) /2.
bin_widths = (bin_edges[1:] - bin_edges[:-1])
# To make error bar plots for the data, take the Poisson uncertainty sqrt(N)
ErrorBar_testing_A = np.sqrt(Histo_testing_A[0])
# ErrorBar_testing_B = np.sqrt(Histo_testing_B[0])
# Draw objects
ax1 = plt.subplot(111)
# Draw solid histograms for the training data
ax1.bar(bin_centers-bin_widths/2.,Histo_training_B[0],facecolor='red',linewidth=0,width=bin_widths,label='B (Train)',alpha=0.5)
ax1.bar(bin_centers-bin_widths/2.,Histo_training_S[0],bottom=Histo_training_B[0],facecolor='blue',linewidth=0,width=bin_widths,label='S (Train)',alpha=0.5)
ff = (1.0*(sum(Histo_training_S[0])+sum(Histo_training_B[0])))/(1.0*sum(Histo_testing_A[0]))
# # Draw error-bar histograms for the testing data
ax1.errorbar(bin_centers, ff*Histo_testing_A[0], yerr=ff*ErrorBar_testing_A, xerr=None, ecolor='black',c='black',fmt='.',label='Test (reweighted)')
# ax1.errorbar(bin_centers, Histo_testing_B[0], yerr=ErrorBar_testing_B, xerr=None, ecolor='red',c='red',fmt='o',label='B (Test)')
# Make a colorful backdrop to show the clasification regions in red and blue
ax1.axvspan(pcut, c_max, color='blue',alpha=0.08)
ax1.axvspan(c_min,pcut, color='red',alpha=0.08)
# Adjust the axis boundaries (just cosmetic)
ax1.axis([c_min, c_max, h_min, h_max])
# Make labels and title
plt.title("Higgs Kaggle Signal-Background Separation")
plt.xlabel("Probability Output (Gradient Boosting)")
plt.ylabel("Counts/Bin")
# Make legend with smalll font
legend = ax1.legend(loc='upper center', shadow=True,ncol=2)
for alabel in legend.get_texts():
alabel.set_fontsize('small')
# Save the result to png
plt.savefig("Sklearn_gbc.png")